Prof. Boan Li's team developed An extraction-free method for rapid detection of CYP2C19 2/3/17 polymorphisms in one tube using melting curve analysis
Single nucleotide polymorphism (SNP) involves one base difference and has important clinical significance. When patients with the same disease take the same drug, they may have different reactions, and the drugs are classified as effective, effective, and ineffective. A single SNP may alter the expression of the protein, thereby altering the drug's target site, resulting in reduced or lack of affinity. Therefore, SNP analysis helps to achieve correct diagnosis and effective treatment.
Some traditional methods for detecting SNPS, such as dideoxy sequencing, pyrosequencing, whole genome sequencing, qPCR, etc., can reveal important features related to SNPS, such as identifying the type and exact location of SNPS. However, these methods are cumbersome and expensive, and are not suitable for general use. Recently, the research team developed a detection strategy based on PCR amplification combined with melt curve analysis, which can simultaneously detect 3 SNP loci CYP2C19 * 2/3/17, a total of 9 genotypes in 1 tube, and achieve rapid detection within 1 h. The research results are entitled "An Extrusion-Free Method for Rapid Detection of CYP2C19 * 2/3/17 Polymorphisms in One Tube using Melting Curve. The study, "Analysis," is published online in the BIOTECHNOLOGY JOURNAL.
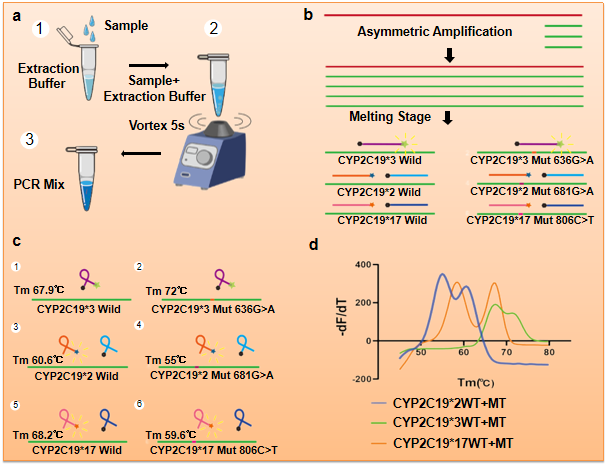
In this study, the initial template was amplified by asymmetric amplification and detected by a combination of adjacent probes and Taqman probes. The 3 'end of the CYP2C19 * 2 probe modifies the FAM fluorophor, the 5' end of the second probe segment adjacent to the 1 base interval modifies the BHQ1 quenching group, and the 3 'end modifies the phosphate group P to block the extension. Similarly, the 3' terminal modified fluorophore of the first probe of CYP2C19 * 17 is ROX, and the quenching group of the second probe is BHQ2. During the study, it was found that the presence of multiple adjacent probes in a tube leads to mutual interference between primers and probes. The method of adjacent probe detection can eventually accommodate two polymorphic sites in a tube. Therefore, we combine the Taqman probe with an adjacent probe. CYP2C19 * 3 is designed as a Taqman probe: the 5 'end is modified by HEX fluorophore, and the 3' end carries a BHQ1 quenching group. Because the binding force between the probe and the template chain decreases with the increase of temperature, the melting peak appears. Because there is a base difference between the homozygous wild-type and mutant samples, the Tm values are different at the melting stage, and the final test results show different Tm values.
In addition, combined with the sample release agent, the sample can be quickly extracted in a few seconds, and the detection system is not limited to blood samples, but also can be applied to oropharyngeal and saliva samples, increasing the diversity and convenience of sampling. We tested 93 clinical blood samples at the same time, verifying that the detection system we established can quickly and accurately identify different genotypes, and the accuracy and effectiveness of the detection was verified by Sanger sequencing. The detection technology based on real-time polymerase amplification combined with melt curve analysis will become a powerful tool to detect and guide the use of clopidogrel because of its advantages of accuracy, speed, simple operation and low cost.
The first author of the paper is Guo Jianguang, a doctoral student in the School of Life Sciences, Xiamen University; You Weixin, a doctoral student, is the co-first author of the paper; and Li Boan, professor in the School of Life Sciences, Xiamen University, and Dr. Rui Zhang, the First Affiliated Hospital of Xiamen University, are the co-corresponding authors of the paper. This project is supported by the National Key Research and Development Plan (2020YFA0112300) and the National Natural Science Foundation (81972458). 82103092), Xiamen Cell Therapy Research Project (3502Z20214001), Fujian Provincial Natural Science Foundation Project (2022J05312), Fujian Provincial Health Education Joint Research Project (2019-WJ-34) and "111 Program" sponsored by the State Administration of Foreign Experts Affairs and the Ministry of Education (B06016).
Paper link:https://doi.org/10.1002/biot.202300207


